Plants of the Caprifoliaceae family are widely cultivated worldwide as ornamental plants owing to their numerous, sweet smelling, beautiful flowers and fruits. Heptacodium miconioides Rehd., a member of the family, is endemic to eastern China and is cultivated as a popular ornamental plant in North America and European countries. It has a rather novel and beautiful trait of high
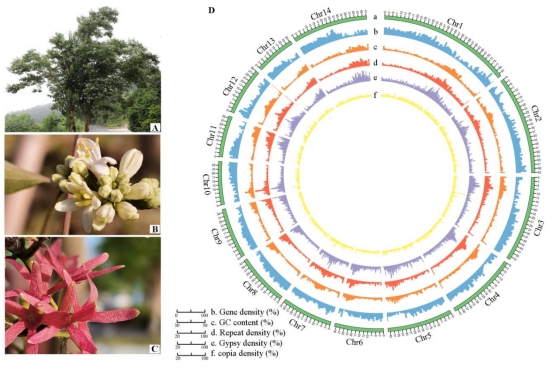
horticultural value, that is, its sepals persist and enlarge, turning purplish red. Here, we report the chromosome-level genome assembly of H. miconioides to understand its evolution and floral characteristics. The 622.28 Mb assembled genome harbored a shared whole-genome duplication with a related species, Lonicera japonica. Comparative genomic analysis suggested that
chromosome fission events following genome duplication underlie the unusual chromosome number of these two species, as well as chromosome fission of another five chromosomes in H. miconioides, giving rise to a haploid chromosome number of 14 (versus 9 in L. japonica). In addition, based on transcriptome and chloroplast genome analysis of 17 representative species in the Caprifoliaceae, we assumed that large structural variations in the chromosomes of H. miconioides were not caused by hybridization. Changes in the candidate genes of the MADS-box family were detected in the H. miconioides genome, including AP1-, AP3-, and SEP- expanded,
which might underlie the sepal elongation and development in this species. The current findings provided a critical resource for genome evolution studies in Caprifoliaceae and it was an example of how multi-omics data can elucidate the regulation of important ornamental traits.